Are you looking for a project for your Bachelor or Master thesis? We are always happy to welcome new team members! Below you can find an overview of the currently available projects per research topic.
Please note: the descriptions are outlines and represent possible directions of the projects during the next two years (2025-2027). The final direction of your project depends on your input and ideas, as well as on available funding and data sources.
Get in touch if you are interested! You can directly contact us or the responsible contact person of each topic.
Cheers,
Anja Westram (anja.m.westram@nord.no) and Joost Raeymaekers (joost.raeymaekers@nord.no)
1. Ecology and evolution of Lake Tanganyika sardines
Contact person: Ivain Martinossi
Collaborators: Joost Raeymaekers, Ilona Strammer, Rashidi Bilali & Leona Milec

Background information
In this project, we focus on two sardine-like freshwater fishes from Lake Tanganyika, Limnothrissa miodon and Stolothrissa tanganicae, which feed millions of people in Central and West Africa. We investigate their ecology, evolution and resilience to climate change and fishing pressure to inform fisheries management and aid the development of sustainable management practices. You can find more info about the project here.
The main goal of this MSc thesis proposal is to document ecological and evolutionary change in the pelagic fish stocks over the last 30 years. This will inform us about the resilience of the lake’s pelagic fisheries to climate change and harvesting.
Brief experimental design/methodology or type of research carried out
- Fieldwork at Lake Tanganyika (DR Congo/Zambia)
- Species identification & dissection
- DNA studies, metabarcoding, stable isotope analysis, or geomorphometrics
2. Reconstructing size distributions and estimating fish stock growth parameters using partial historical samples from Lake Tanganyika
Contact person: Ivain Martinossi
Collaborators: Joost Raeymaekers, Ilona Strammer & Rashidi Bilali

Background information
Lake Tanganyika harbours important fish stocks, which may be affected by ongoing climate change. To understand how fish stocks are changing over time, it is crucial to have access to historical samples. But sometimes, they are not collected or preserved in the best way! Can we be clever enough to use partial samples (fish tails) to reconstruct the size distribution of the population and determine important demographic parameters of the stock? These could then be compared to data that is being sampled currently.
Brief experimental design/methodology or type of research carried out
- Process and photograph historical samples
- Figure out a way to estimate full body size from partial sample
- Learn data analysis and visualisation
- Learn how to estimate growth parameters of fish stocks
- Assess the evolution of the stock over time
3. Genetics of scavenging amphipods in sub-Arctic fjords
Contact person: Irina Smolina
Collaborators: Joost Raeymaekers and Henning Reiss
Background information
Scavenging amphipods are among the most dominant group in deep-sea environments and plays an important role in carbon cycling. The knowledge on taxonomic and genetic diversity of amphipods is still limited in sub-Arctic fjord systems. Our first pilot barcoding data hints at genetic differentiation between Skjerstadfjord and Saltfjorden for amphipods Tmetonix cicada, as well as 2 distinct clades within Saltfjorden. More individuals and locations need to be studied to illuminate the genetic differentiation in this species in sub-Arctic fjord systems.
Brief experimental design or type of research carried out
- Field work on collecting new amphipods from neighbouring fjords. Some previous samples are also available.
- Wet lab: morphological species identification, DNA extraction and mitogene barcoding. Optional, advanced library preparation and sequencing using Oxford Nanopore technology.
- Dry lab: genetic data analysis and possibly for advanced bioinformatic analysis of high-throughput sequencing data
4. Epizoic diatoms as health indicators of sea turtles
Contact person: Roksana Majewska
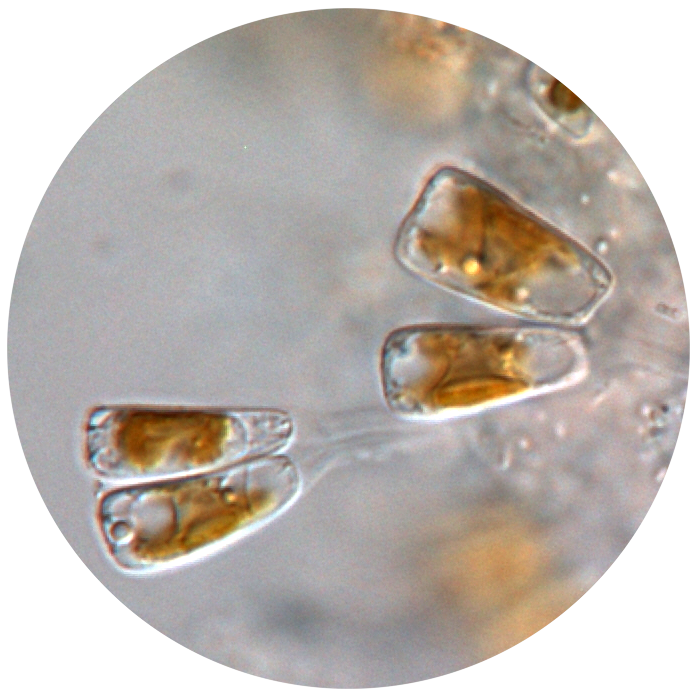
Background information
Diatoms, single-celled algae, are one of the most successful, abundant and species-rich groups of primary producers in the world. Although diatoms grow on a wide range of submerged surfaces, some diatom species and even entire genera are highly specialised and thrive exclusively on the surface of glabrous aquatic animals (such as sea turtles). Although the nature of this peculiar association is largely unknown, the survival of the epizoic diatoms seems to depend entirely on the health and wellbeing of their host.
Brief experimental design/research carried out
- biofilm sample processing and permanent slide preparation
- diatom identification and community composition analysis (microscopy and molecular approaches: metabarcoding)
- statistical analyses of community abundance data
- bioindication using diatoms
5. What do nature managers and conservation practitioners think about genetic diversity?
Contact person: Anja Westram
Collaborators: Joost Raeymaekers

Background information
Genetic diversity within species represents an important component of biodiversity. It needs to be conserved if we want to maintain functional natural populations & ecosystem services. But do conservation practitioners have the necessary interest and knowledge about genetic diversity?
Brief experimental design/type of research carried out
In this MSc project, you will help us to identify local nature managers and conservation practitioners in Norway, and design and conduct a survey about their interest in and knowledge about genetic diversity. You will analyse the data with statistical analyses and formulate recommendations for policy.

6. A population genomic comparison of two stickleback fishes
Contact person: Thijs Mattheus Peter Bal
Collaborators: Joost Raeymaekers and Arun Gowda

Background information
Populations of the three-spined stickleback show stronger signs of adaptation than populations of the nine-spined stickleback. Therefore these species are excellent models to study why some species evolve more than others, even though they live in the same landscape.
Brief experimental design/type of research carried out
Using population genomic and bioinformatic tools on existing whole-genome sequencing data, we will test how structural variants (CNVs, indels, duplications and inversions) are distributed in the genome, and analyse which of these structural variants are the target of natural selection.