Are you looking for a project for your Bachelor or Master thesis? We are always happy to welcome new team members! Below you can find an overview of the currently available projects per research topic.
Please note: the descriptions are outlines and represent possible directions of the projects during the next two years (2022-2024). The final direction of your project depends on your input and ideas, as well as on available funding and data sources.
Get in touch if you are interested! You can directly contact us or the responsible contact person of each topic.
Cheers,
Anja Westram (anja.m.westram@nord.no) and Joost Raeymaekers (joost.raeymaekers@nord.no)
1. Parallel evolution in a marine snail
Contact person: Anja Westram
Collaborators: Ana Peris, Bingqian Han, Joost Raeymaekers
Topic 1A: The role of a chromosomal inversion in temperature adaptation in marine snails
 How do organisms adapt to their local environment? This question is a main focus of evolutionary biology. It becomes especially urgent in times of anthropogenic climate change. The marine snails Littorina saxatilis and Littorina arcana live on rocky shores and is represent an excellent system to study adaptation to spatial temperature gradients. We are interested in how adaptation works at both the phenotypic and the genomic level. In previous work on L. saxatilis, we have found that a chromosomal inversion (a large mutation that flips a part of the chromosome) is associated with adaptation to high shore levels, where exposure to high temperatures is more frequent and snails are more likely to dry out.
How do organisms adapt to their local environment? This question is a main focus of evolutionary biology. It becomes especially urgent in times of anthropogenic climate change. The marine snails Littorina saxatilis and Littorina arcana live on rocky shores and is represent an excellent system to study adaptation to spatial temperature gradients. We are interested in how adaptation works at both the phenotypic and the genomic level. In previous work on L. saxatilis, we have found that a chromosomal inversion (a large mutation that flips a part of the chromosome) is associated with adaptation to high shore levels, where exposure to high temperatures is more frequent and snails are more likely to dry out.
In this project, we will test how this inversion contributes to adaptation to different microhabitats, and if it contributes to adaptation in both species (L. saxatilis and L. arcana). We will sample snails from different habitats on the coast near Bodø. We will then perform physiological tests for temperature adaptation in the lab and identify the species via dissection and with genetic markers. We will also genotype genetic markers in the inversion to identify the karyotype of each individual. We will then perform statistical analyses to test for associations between karyotype, physiological adaptation and habitat, and for differences between the two species.
This project will be interesting for a student interested in evolutionary biology (selection and adaptation), who would like to combine fieldwork, wet lab work, genetic data analysis and statistical analyses. The student will gain skills in experimental design, field- and labwork, and statistics. Creativity and own ideas are expected. The student will closely collaborate with the Littorina team, working on questions directly related to this project.
Topic 1B: Speciation in marine snails
 Speciation is a crucial and fascinating process in evolutionary biology. Speciation occurs when populations become reproductively isolated, i.e. lose their ability to exchange genes. The marine snails Littorina saxatilis and Littorina arcana are closely related and morphologically extremely similar – what limits their ability to exchange genes?
Speciation is a crucial and fascinating process in evolutionary biology. Speciation occurs when populations become reproductively isolated, i.e. lose their ability to exchange genes. The marine snails Littorina saxatilis and Littorina arcana are closely related and morphologically extremely similar – what limits their ability to exchange genes?
In this project, we will analyse reproductive barriers between these two species by asking whether they mate in the field and testing whether they can sometimes produce hybrid offspring. We will collect mating pairs of snails from the coast near Bodø, sampling different habitats and areas where the two species differ in relative frequency. We will then use genetic markers to identify the species (due to their morphological similarities, morphological identification is not reliable). In addition, we will analyse the embryos the female snails carry, in order to detect whether there is any evidence for hybrids between the two species. With these data, we will be able to test whether the two species are isolated via 1) mating isolation (and whether this depends on the environment or relative frequency of the species), and 2) the inability to produce viable hybrids.
This project will be interesting for a student interested in evolutionary biology (speciation), who would like to combine fieldwork, wet lab work, genetic data analysis and statistical analyses. The student will gain skills in experimental design, field- and labwork, and statistical analyses. Creativity and own ideas are expected. The student will closely collaborate with the Littorina team and will be involved in international collaborations.
2. Ecology and evolution of Lake Tanganyika sardines
Contact person: Leona Milec
Collaborators: Joost Raeymaekers
 In this project, we focus on two sardine-like freshwater fishes from Lake Tanganyika, Limnothrissa miodon and Stolothrissa tanganicae, which feed millions of people in Central and West Africa. We investigate their ecology, evolution and resilience to climate change and fishing pressure to inform fisheries management and aid the development of sustainable management practices. Here you can find more info about the project.
In this project, we focus on two sardine-like freshwater fishes from Lake Tanganyika, Limnothrissa miodon and Stolothrissa tanganicae, which feed millions of people in Central and West Africa. We investigate their ecology, evolution and resilience to climate change and fishing pressure to inform fisheries management and aid the development of sustainable management practices. Here you can find more info about the project.
Topic 2A: Adaptation to new lake environments following introduction/invasion – the case of L. miodon
 The freshwater sardine L. miodon, originally endemic to Lake Tanganyika, has been introduced into several smaller lakes in the surrounding countries for fishery purposes, including the natural Lake Kivu and the man-made reservoirs Kariba and Cahora Bassa. Using RAD-tag sequencing data and a combination of population genomic and demographic modelling approaches, we aim to address how the sudden exposure of L. miodon to its new and distinct environments may have influenced its life history, genetic diversity and population differentiation. Knowledge of phenotypic and genetic changes following introduction will prove instrumental to meet the unique management needs of each lake. In addition, we are interested in changes in diet and gut microbial communities following invasion. The sardines may have undergone diet changes since prey communities may differ between Lake Tanganyika and the new lakes. Gut microbiota co-evolve with their host and play a crucial role in the host’s functioning of various biological processes, but changes may occur upon the establishment of new ecological niches. The student will closely collaborate with Leona to participate in laboratory work, and genetic and bioinformatic analyses.
The freshwater sardine L. miodon, originally endemic to Lake Tanganyika, has been introduced into several smaller lakes in the surrounding countries for fishery purposes, including the natural Lake Kivu and the man-made reservoirs Kariba and Cahora Bassa. Using RAD-tag sequencing data and a combination of population genomic and demographic modelling approaches, we aim to address how the sudden exposure of L. miodon to its new and distinct environments may have influenced its life history, genetic diversity and population differentiation. Knowledge of phenotypic and genetic changes following introduction will prove instrumental to meet the unique management needs of each lake. In addition, we are interested in changes in diet and gut microbial communities following invasion. The sardines may have undergone diet changes since prey communities may differ between Lake Tanganyika and the new lakes. Gut microbiota co-evolve with their host and play a crucial role in the host’s functioning of various biological processes, but changes may occur upon the establishment of new ecological niches. The student will closely collaborate with Leona to participate in laboratory work, and genetic and bioinformatic analyses.
Topic 2B: Larval species composition at Lake Tanganyika’s spawning grounds
 The population dynamics of fish are often strongly coupled to their spawning and nursing grounds. Both climate change and fishing pressure in and around these areas are likely to induce changes in the distribution of these grounds and in recruitment success. Juveniles of the semi-littoral clupeid L. miodon, and possibly of the pelagic clupeid S. tanganicae, are heavily harvested in the littoral zone of Lake Tanganyika. However, how much each species makes use of these spawning and nursing grounds remains uncertain. We use COI barcoding and morphometry to identify the larvae, and determine the variation in larval species composition along the North-South axis of the lake. The student will closely collaborate with Leona to participate in field work, laboratory work, and genetic and bioinformatic analyses.
The population dynamics of fish are often strongly coupled to their spawning and nursing grounds. Both climate change and fishing pressure in and around these areas are likely to induce changes in the distribution of these grounds and in recruitment success. Juveniles of the semi-littoral clupeid L. miodon, and possibly of the pelagic clupeid S. tanganicae, are heavily harvested in the littoral zone of Lake Tanganyika. However, how much each species makes use of these spawning and nursing grounds remains uncertain. We use COI barcoding and morphometry to identify the larvae, and determine the variation in larval species composition along the North-South axis of the lake. The student will closely collaborate with Leona to participate in field work, laboratory work, and genetic and bioinformatic analyses.
3. Adaptation to human-impacted environments in stickleback fishes
Contact person: Brijesh Yadav/Aruna M Shankregowda
Collaborators: Joost Raeymaekers and Deepti Patel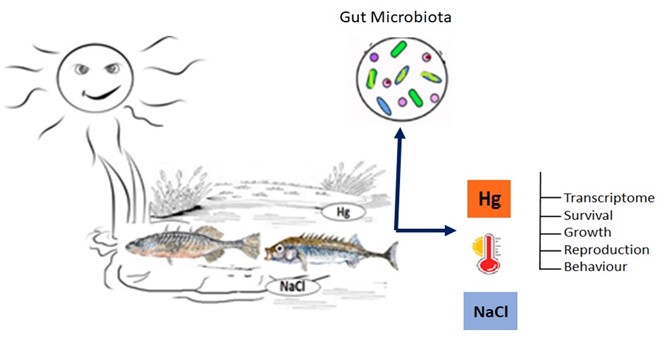
Human-induced pollution features among the greatest challenges that organisms face for survival and adaptation. Aquatic ecosystems are exposed worldwide to varying degrees of pollution. The fitness of their communities and populations has been affected to such an extent that biodiversity is compromised. The adaptive capacity of stickleback fishes, from their cells to their overall performance, allows them to adjust to environmental change. Through a set of field studies on wild stickleback and experiments on lab-reared sticklebacks, we aim to understand their adaptive responses to pollution. This subject will give you an understanding of how organisms adjust to new environmental conditions (through physiological responses or interaction with the microbiome) or adapt to their environment (through evolutionary change).
Topic 3A: Biological responses of three-spined stickleback to mercury and temperature stress
 Mercury (Hg) is one of the most toxic and wide-spread pollutants in aquatic environments. It bio-accumulates as methylmercury (MeHg) in organisms and biomagnifies in the food chain. The rapidly changing climate may enhance these harmful effects because higher temperatures may increase Hg methylation rates. This risk is particularly high in high latitude populations, as those are expected to undergo the most severe temperature changes. We investigate the combined effect of temperature stress and exposure to Hg and use RNA-sequencing to identify differentially expressed genes, proteins, and pathway enrichment. We will use experimental populations as well as populations from pristine and Hg polluted sites. The experimental part will also include the study of Hg accumulation, survival, growth, body condition, and behaviour. The student will closely collaborate with the stickleback team to participate in field sampling, experimental crosses, molecular lab work, and bioinformatic analyses.
Mercury (Hg) is one of the most toxic and wide-spread pollutants in aquatic environments. It bio-accumulates as methylmercury (MeHg) in organisms and biomagnifies in the food chain. The rapidly changing climate may enhance these harmful effects because higher temperatures may increase Hg methylation rates. This risk is particularly high in high latitude populations, as those are expected to undergo the most severe temperature changes. We investigate the combined effect of temperature stress and exposure to Hg and use RNA-sequencing to identify differentially expressed genes, proteins, and pathway enrichment. We will use experimental populations as well as populations from pristine and Hg polluted sites. The experimental part will also include the study of Hg accumulation, survival, growth, body condition, and behaviour. The student will closely collaborate with the stickleback team to participate in field sampling, experimental crosses, molecular lab work, and bioinformatic analyses.
Topic 3B: A study on the impact of mercury in Norwegian lakes
In Scandinavia, toxic mercury (Hg) levels in fish tissue have been steadily declining over the last 50 years (1965-2015), while still remaining high. Hg levels are expected to drop with latitude due to decreased atmospheric deposition. Yet, observations only show a weak effect, presumably because of higher Hg biomagnification rates in subarctic lakes compared to lakes further south. The three-spined stickleback is a commonly used species in ecotoxicological studies and is widely distributed in the Norwegian aquatic ecosystem. In this study, we investigate the impact of mercury on the abiotic and biotic environment of 21 lakes in northern Norway. We collect sticklebacks, water, sediment, benthos, and zooplankton samples to determine mercury concentrations and toxicity levels throughout the lake’s food web. The student will closely collaborate with the stickleback team to participate in field sampling, laboratory work, and data analysis.
4. Evolutionary ecology of two coexisting stickleback species
Contact person: Thijs Mattheus Peter Bal
Collaborators: Joost Raeymaekers and Kostas Sagonas
 Studying the mechanisms behind population divergence and local adaptation is a large aspect of modern evolutionary biology. Increasing our understanding of these concepts is important for conservation ecology and landscape management. It is for instance an important question which evolutionary trajectories different species may take when they experience environmental change. One approach to answer this question is to use natural populations and study how variation is distributed in the system in relation to the landscape.
Studying the mechanisms behind population divergence and local adaptation is a large aspect of modern evolutionary biology. Increasing our understanding of these concepts is important for conservation ecology and landscape management. It is for instance an important question which evolutionary trajectories different species may take when they experience environmental change. One approach to answer this question is to use natural populations and study how variation is distributed in the system in relation to the landscape.
In our research we use ecologically similar and coexisting three-spined and nine-spined stickleback fishes as models for understanding evolutionary change in a natural system. These two species co-occur in the riverine landscape of Belgium and the Netherlands and this system encompasses significant environmental differences on a relatively small geographical scale. We use a broad approach and we utilize different types of data to study the (adaptive) variation between individuals, populations and species.
Topic 4A: The role of genomic structural variants in local adaptation
Single‐nucleotide polymorphisms (SNPs) are one of the most widely used types of genetic variation for studying signatures of local adaptation, but in recent years there has been an increasing interest in the role of structural variants. The main goal of this thesis topic is to identify genomic structural variants that potentially play a role in local adaptation in coexisting three-spined and nine-spined stickleback. Earlier research within this study system using SNPs has revealed stronger signatures of selection in three-spined stickleback than in nine-spined stickleback. However, it is still largely unknown how much structural variants contribute to the level of genetic variation, and more importantly, the role they play in local adaptation in the two species. It is for example possible that three-spined stickleback again show stronger signatures of selection than nine-spined stickleback when looking at this type of genetic variation, strengthening the hypothesis that three-spined stickleback overall have a stronger evolutionary response. Another interesting possibility is that nine-spined stickleback actually show stronger signatures of selection for this type of genetic variation, revealing that selection simply has the potential to leave different genomic signatures of adaptation in species experiencing similar selection pressures. Indeed, all possible findings have major implications for understanding local adaptation in this system and hence it is highly important that the role of genomic structural variants is assessed.
In this project the student gets access to whole genome sequences of 192 individuals of each species. This project is suited for students with a strong interest in (population) genomics and (learning) bioinformatics. There is a lot of data available and different approaches as well as different specific research questions can be explored. For this reason, personal input and creativity by the student is also encouraged. The student will closely collaborate with Thijs and has the chance to be involved in the writing of a manuscript aimed for publication in a peer-reviewed scientific journal.
5. Diversity of micro-organisms in Northern Norway
Contact person: Lynn Govaert
Collaborators: Joost Raeymaekers
Are you fascinated by small ponds and lakes? Did you ever wonder what microscopic life these freshwaters harbour? If yes, then we might have a thesis topic for you! In the freshwater ciliate team, we investigate the species diversity and genetic diversity of ciliates in Norway. Ciliates belong to the protists, which are eukaryotes that are neither plants, animals or fungi, but which for instance also includes algae and slime molds. One of the most famous protists is the ciliate Paramecium caudatum (tøffeldyr in Norwegian or pantoffeldier in Dutch), which you might have encountered when you were a child and for the first time looked through a microscope. In this project, you will survey protozoan communities of natural freshwater ponds along an latitudinal gradient in Norway. In particular, you will identify the different species found via metabarcoding and microscopy. This project will contribute to the largely unknown micro-organismal diversity harboured in freshwater ponds in Northern Norway. Depending on your interest, you can also include more experimental or molecular lab work in your thesis. For more information about the freshwater ciliate team, please check my webpage here or here.
6. Molecular evolution of skuas, predatory sea birds
Contact person: Irina Smolina
Collaborators: Kirstin Janssen (UiT Tromsø) and Truls Moum
#birds #phylogeny #bioinformatics #molecular biology #evolution
The skuas are a small group of predatory seabirds with intriguing questions related to their ecology and evolution. Skua taxonomy is still under discussion: should all species belong to one genus Stercorarius or should they be split into two distinct groups (genera Stercorarius and Catharacta)? One species (pomarine skua) is intermediate between the two groups. Is the intermediate species a result of hybridisation and/or selection acting on morphological traits?
We have samples of most skua species for genetic analyses to explore the molecular evolution and relationships of their mitochondrial and nuclear genes.


Left: Arctic skua (Photo: Tycho Anker-Nilssen). Right: Great skua (Photo: Sebastien Descamps).
Objectives: to explore the molecular evolution and phylogeny of skuas based on mitochondrial and nuclear DNA sequence data using various bioinformatics methods. Further specific goals and project direction will depend on interest, motivation and skills of the candidate.
Available resources: whole genome sequences of 7 individuals representing 6 species, reference nuclear genome and mitogenomes from NCBI, population tissue samples.
Proposed tasks:
- Nuclear and mitochondrial genome assemblies.
- Phylogenetic analyses based on mitochondrial and nuclear DNA sequence data.
- (Optional) Exploration of the molecular evolution of target nuclear genes/gene families.
- (Optional) Investigation of possible hybridisation, selection, genetic species boundaries.
- (Optional) Exploration of mitochondrial sequence duplications and their role.
Approach: mostly bioinformatics, molecular lab work could be added.
Expected general skills to be obtained after completion:
- operation and manipulation of NGS data, working in command line, scripting, experience with various common bioinformatics software
- objective assessment of current state of the topic and identification of knowledge gap, efficient written and oral communication and presentation of performed work
- (optional) ability to perform typical molecular biology workflow from DNA extraction to library preparation and Illumina sequencing.